

There are two approaches you can use to input data for prediction shown in Figure 1.
1. Paste FASTA-format text in the area circled with the red line
2. Please upload protein sequence(s) in FASTA from your local disk via the button marked with the blue line.

Figure 1. GUI of sequence(s) submission
In general, the time of computation won't be too long, and you can wait in front of your screen until the prediction result is available. Otherwise, you can leave a valid e-mail address, and the system will send a mail to inform you when the result has come out. (Figure 2)
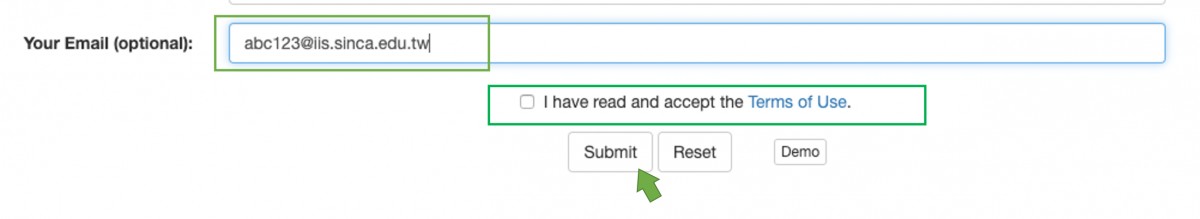
Figure 2. key-in your e-mail and check the box of "Terms of Use" to complete the submission with e-mail notification.
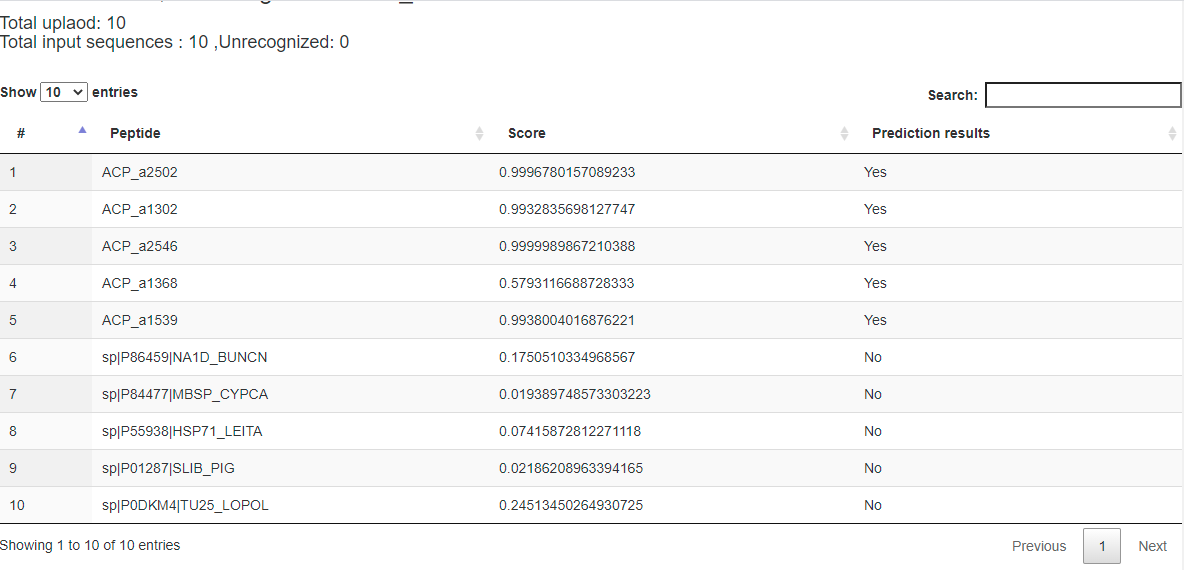
Figure 3. Prediction results of the demo FASTA file.
The column "Peptide" lists the names of input peptides. The column "Score" shows prediction scores indicating how much probability that a peptide contains ACP activity. The column "Prediction results" shows whether the peptide is an ACP. Here, the threshold of prediction score is around 0.472. "YES" means the prediction score of the peptide more than 0.472; otherwise, "No" says the prediction score of the peptide less than 0.472. (Figure 3)
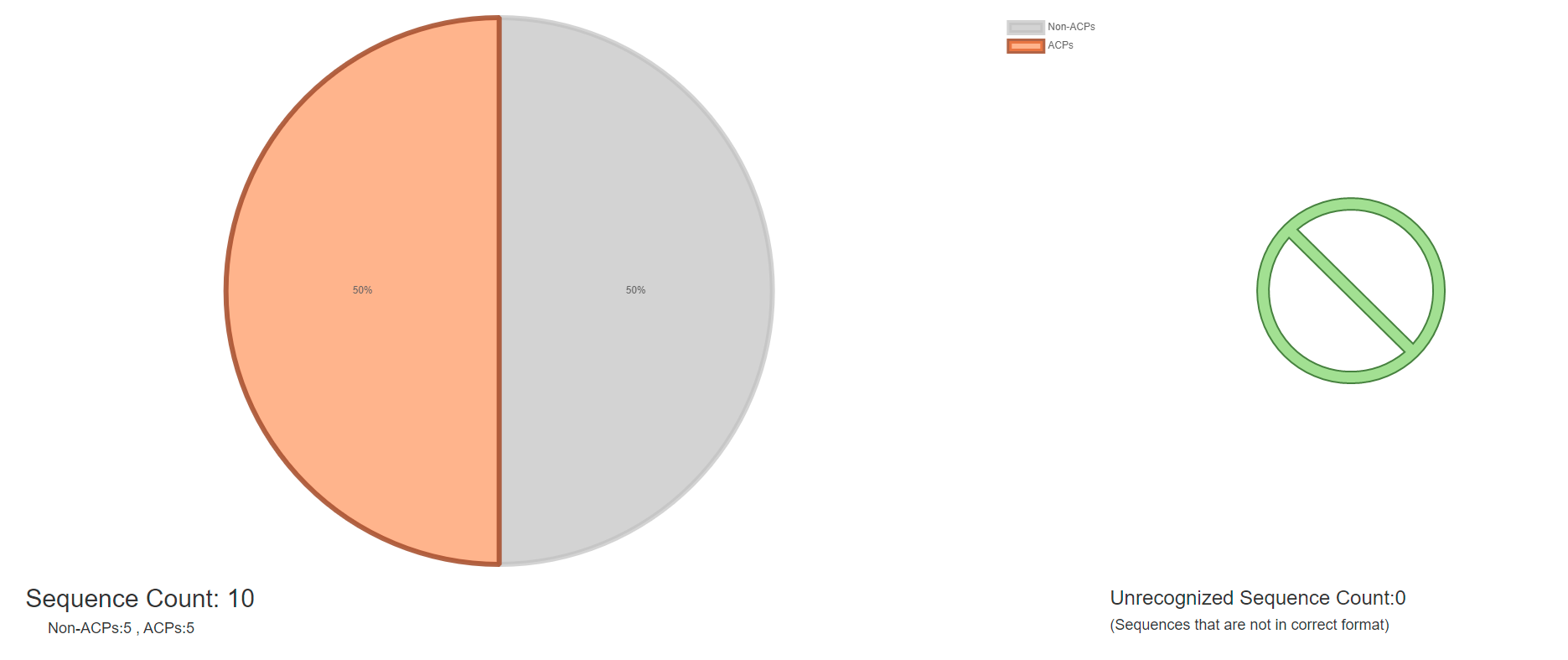
Figure 4. Visualization of ACP prediction with statistics.
There is a total of 10 peptides in the demo FASTA file, and the prediction result shows that it contains 5 ACPs and 5 Non- ACP. unrecognized sequence also visualized and counted which listed in the input data.
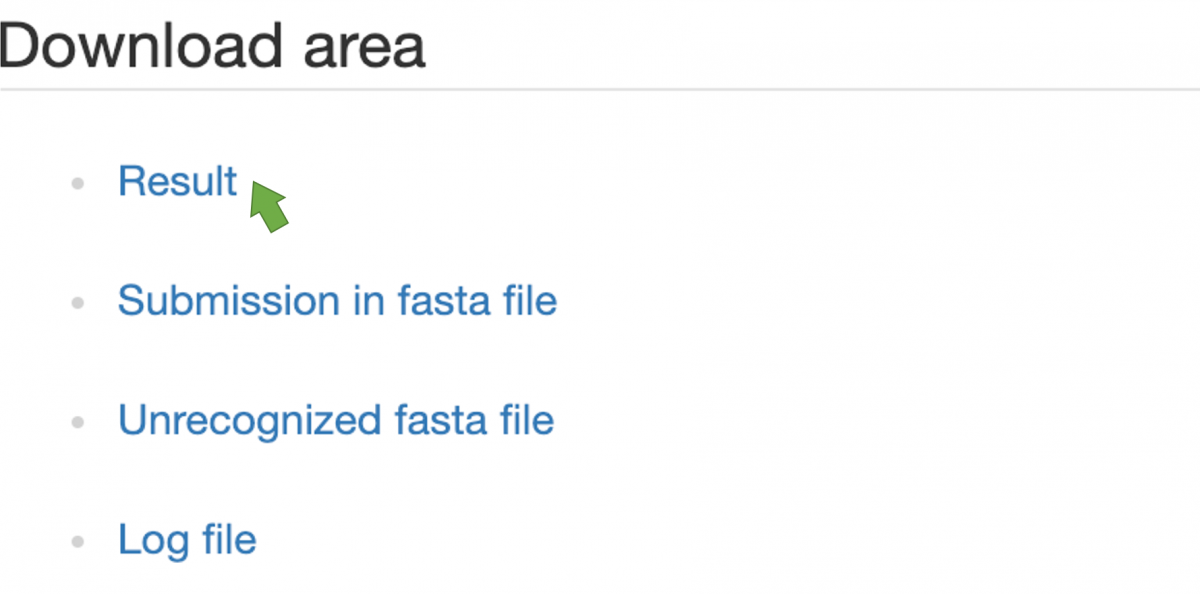
Figure 5. Available files listed in Download area
Users can download their prediction results in the "Download area" (Figure 5). Click "Result" to download prediction results represented in CSV. Here are also "Submission in fasta file," "Unrecognized fasta file," and even "Log file" provided as references.
1. ACPs data for model tuning [download] (1912)
2. Non-ACPs data for model tuning [download] (1912)
3. ACPs data for testing [download] (212)
4. Non-ACPs data for testing[download] (212)
5. ACPs data for final model training [download] (2124)
6. Non-ACPs data for final model training [download] (2124)
Copyright © 2021 Institute of Information Science, Academia Sinica, TAIWAN. |
All Rights reserved. |