

There are two approaches you can use to input data for prediction shown in Figure 1.
1. Paste FASTA-format text in the area circled with the red line
2. Please upload protein sequence(s) in FASTA from your local disk via the button marked with the blue line.
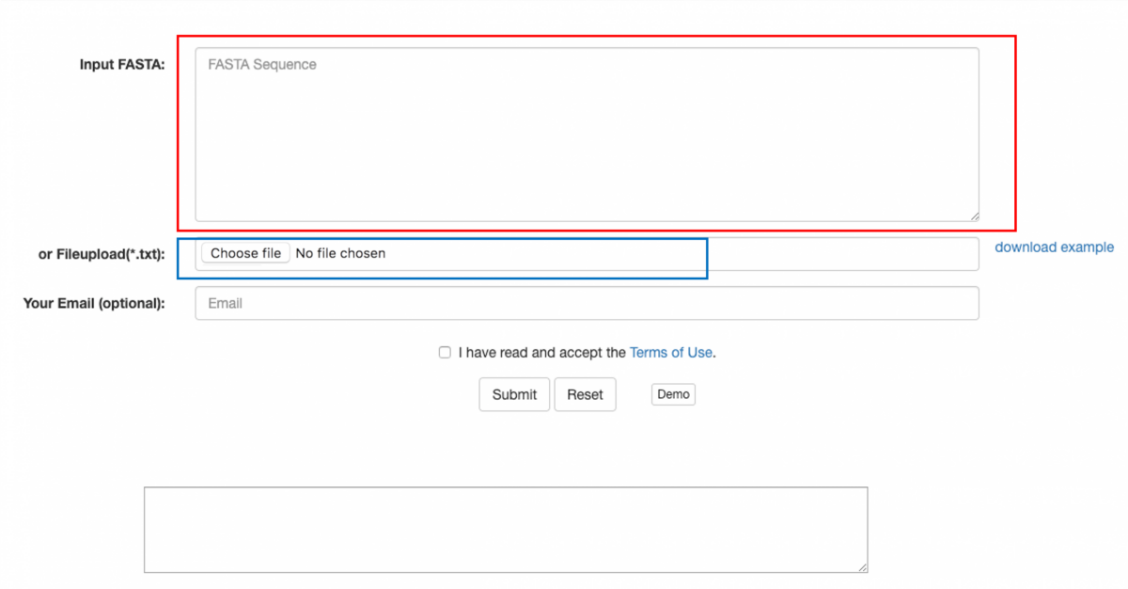
Figure 1. GUI of sequence(s) submission
In general, the time of computation won't be too long, and you can wait in front of your screen until the prediction result is available. Otherwise, you can leave a valid e-mail address, and the system will send a mail to inform you when the result has come out. (Figure 2)
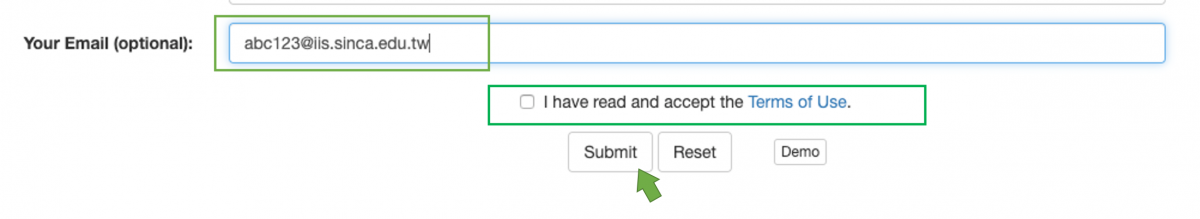
Figure 2. key-in your e-mail and check the box of "Terms of Use" to complete the submission with e-mail notification.
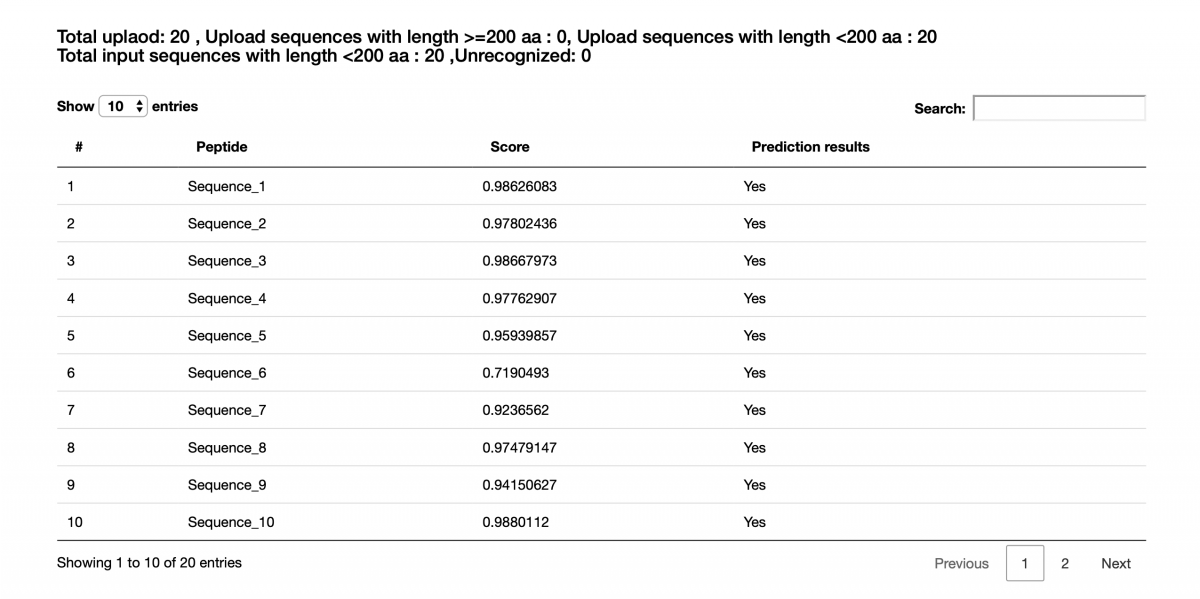
Figure 3. Prediction results of the demo FASTA file.
The column "Peptide" lists the names of input peptides. The column "Score" shows prediction scores indicating how much probability that a peptide contains AMP activity. The column "Prediction results" shows whether the peptide is an AMP. Here, the threshold of prediction score is around 0.5. "YES" means the prediction score of the peptide more than 0.5; otherwise, "No" says the prediction score of the peptide less than 0.5. (Figure 3)

Figure 4. Visualization of AMP prediction with statistics.
There is a total of 20 peptides in the demo FASTA file, and the prediction result shows that it contains 19 AMPs and 1 Non- AMP. Long peptide and unrecognized sequence also visualized and counted which listed in the input data.
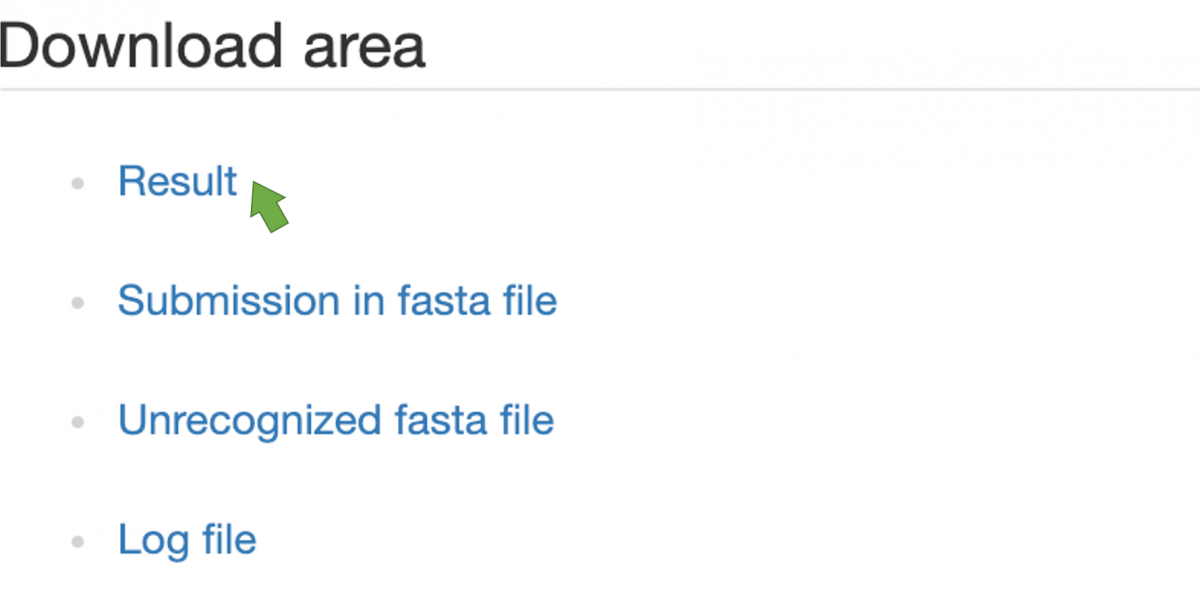
Figure 5. Available files listed in Download area
Users can download their prediction results in the "Download area" (Figure 5). Click "Result" to download prediction results represented in CSV. Here are also "Submission in fasta file," "Unrecognized fasta file," and even "Log file" provided as references.
Our AMPs dataset was obtained from four databases: APD3 (Li et al., 2015), LAMP (Zhao et al., 2013), CAMP3 (Thomas et al., 2010), and DRAMP (Kang et al., 2019). We downloaded all anti-bacterial AMP data from the four databases and excluded AMPs with sequence lengths shorter than ten amino acids and those containing unusual amino acids. Also, duplicate data were be removed. The non-AMP dataset is a combination of real-world peptides and artificially generated sequences. Real-world peptides were obtained from UniProt with the following criteria: (1) sequence length between 10 to 50, (2) without AMP-related keywords in annotation, such as 'Antimicrobial,' 'Antibiotic,' 'Amphibian defense peptide,' and 'Antiviral protein.' Artificially generated sequences were randomly generated from 20 essential amino acids, with the same length distribution as the AMP dataset.
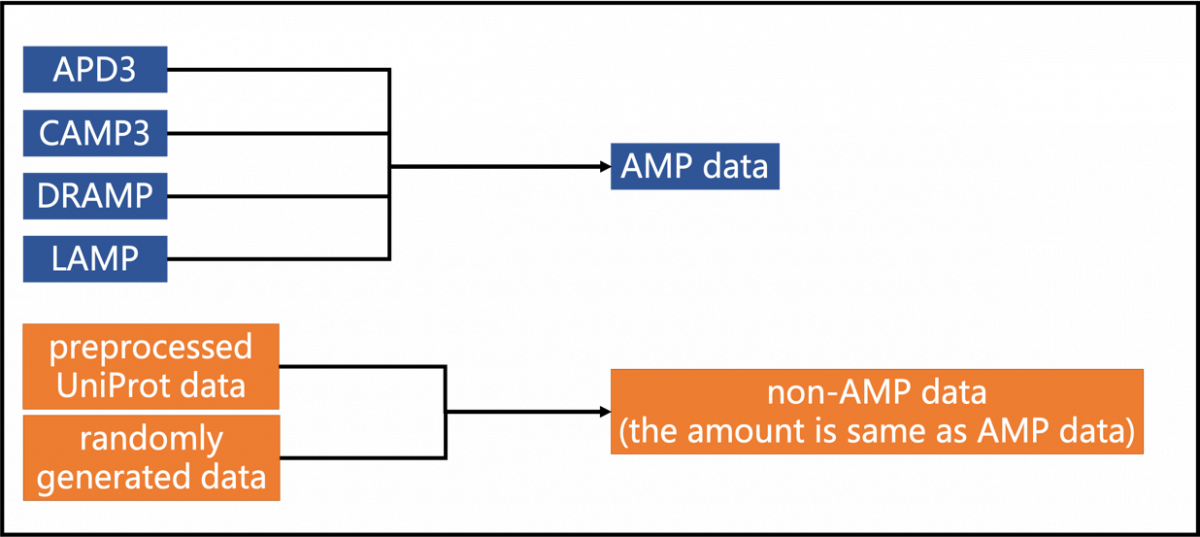
Figure 6. Data used to build the final model for AI4AMP website.
We used 3528 AMP sequences and 3528 non-AMP sequences to construct and tune our model. AMP data was obtained from the original collection of the AMP dataset, excluding the data from the LAMP database and sequences with high identity to this set (> 90%). The non-AMP data, 1778 sequences were from the dataset used in Antimicrobial Peptide Scanner vr.2 (Veltri et al.), appended with 1750 data randomly generated sequences.
The external AMP testing dataset was composed of 565 AMPs collected from the
LAMP database by excluding sequences sharing 90% or higher sequence identity and 565 non-AMP sequences (randomly selected from non-AMP short peptides collected from UniProt plus randomly generated sequence with the same peptide length distribution to the collected AMPs).
1. AMPs data for model tuning [download] (3528)
2. Non-AMPs data for model tuning [download] (3528)
3. AMPs data for external testing [download] (565)
4. Non-AMPs data for external testing[download] (565)
5. AMPs data for final model training [download] (6623)
6. Non-AMPs data for final model training [download] (6623)
Copyright © 2021 Institute of Information Science, Academia Sinica, TAIWAN. |
All Rights reserved. |