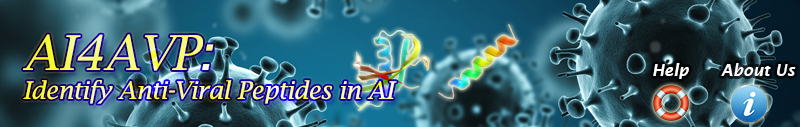



There are two methods used to input sequence(s) for prediction (Figure.1):
•We ONLY accept FASTA-format text
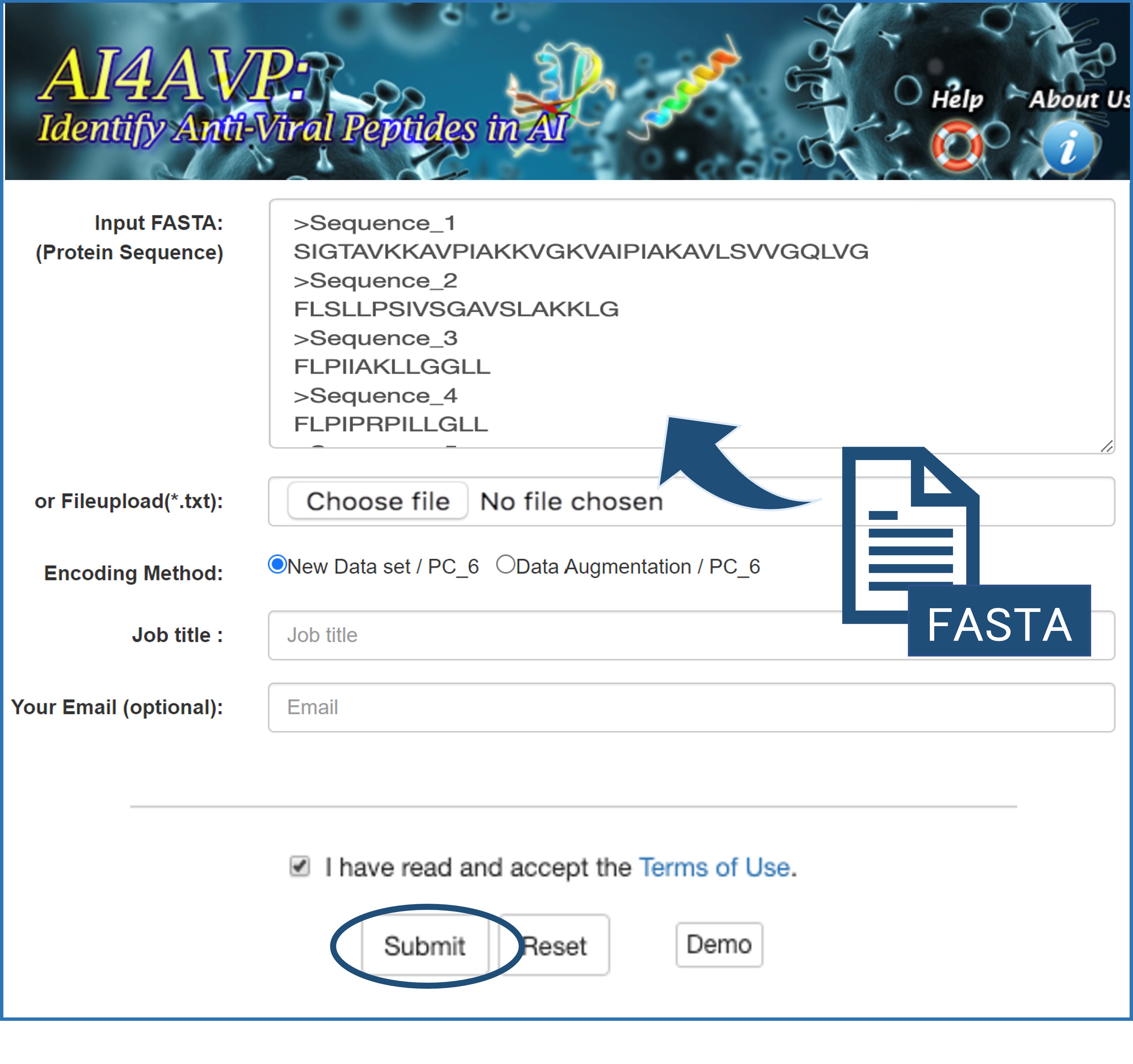
Figure 1. GUI of AI4AVP.
•Usually, the time used for computation will not be too long. The user may wait until the result is available or leave a valid email address; the system will email the submitter when the result has come out.
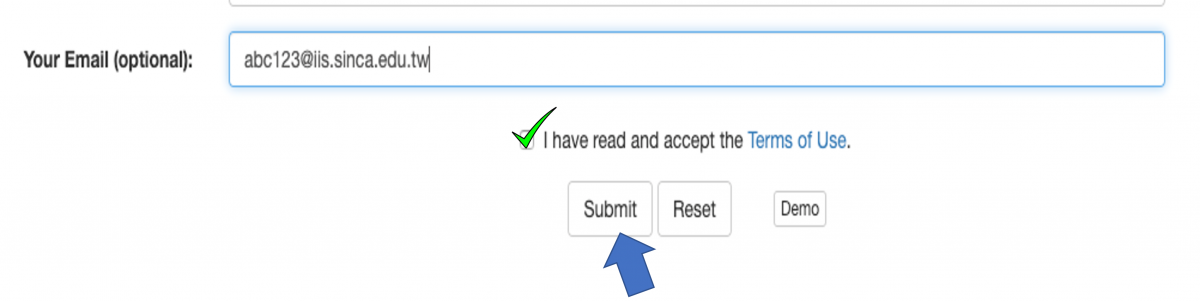
Figure 2. key-in a validated email and read the "Terms of Use" if needs.
Figure 3. The prediction results of the demo FASTA file.
The figure above shows the prediction results of the demo FASTA file. The column "Peptide" lists the names of input peptides. The column "Score" shows prediction scores indicating how much probability a peptide contains AVP activity. The column "Prediction results" shows whether the peptide is an AVP or not. Here, the threshold is a 0.5 prediction score. "YES" means the prediction score of the peptide is more than 0.5. Otherwise, "No" means the prediction score of the peptide is less than 0.5.
Meanwhile, for those submitted sequences with the length >= 200 a.a., AI4AVP will cut the sequence(s) into several ones by a sliding window (windows size =200 aa and step size =50 aa).

Figure 4. Overall the result of demo submission in piecharts.
The pie charts above show the visualization of the results in a global view (Figure 4). There are a total of 20 peptides in the demo FASTA file, and the prediction result shows that it contains 11 AVPs and 8 Non- AVPs. Submitted long peptides (>200 aa) and unrecognized sequences are visualized and counted in separated piecharts. However, it must be emphasized that most AVPs composed of essential amino acids with a length shorter than 50 amino acids.
Figure 5. The download area for each submission.
Users can download their prediction results in the "Download area." Click "Result" to download the prediction results represented in the CSV file. Here are also "Submission in fasta file," "Unrecognized fasta file," and "Logfile" provided as references.
|
2012 Dataset |
[link] (506 positive and negative data points) |
|
New dataset |
|
|
Positive Dataset(Training + Validation): |
real AVP [download] (2,641) |
|
Negative Dataset (Training + Validation): |
Non-AVP [download] (16,995) |
|
GAN dataset: |
AVP [download] in GAN (14,354) |
|
External dataset for Validation: |
|
|
|
Testing AVP [download] (293) |
|
|
Testing Non_AVP [download] (293) |


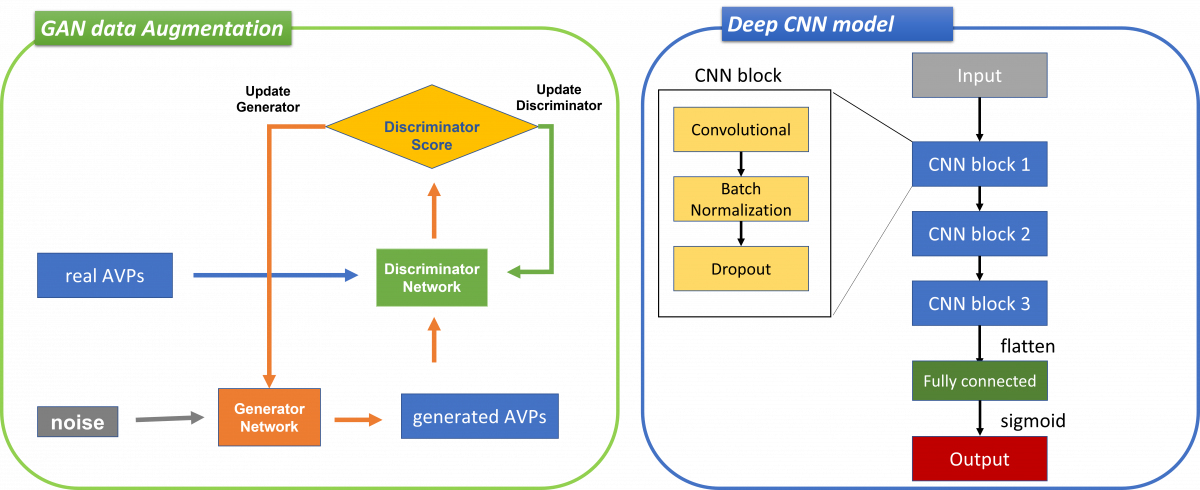
Figure A1. The structure of models for GAN and Deep CNN used in this study.
The model used for AI4AVP was proposed in our study, 'Developing an Antiviral Peptides Predictor with Generative Adversarial Network Data Augmentation' (BioRxiv, 2021).' We used a generative adversarial network (GAN) to generate a positive training dataset to allow the deep CNN classifier to train on balanced datasets. Then, we encoded peptides using the PC6 protein-encoding method and trained the AI4AVP model based on three CNN blocks.
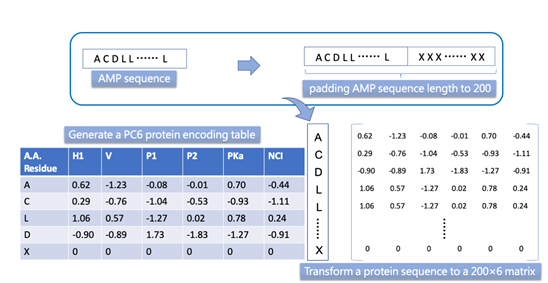
Figure A2. Encoding the amino acids sequence according six physicochemical properties (PC6).
The idea of the PC6 encoding method is to use physicochemical properties as word embeddings. Each amino acid character in sequence would be replaced with a vector composed of six physicochemical property values (PC6). The six features of PC6 include hydrophobicity (H1), the volume of side chains (V), polarity (Pl), and pH at the isoelectric point (pI), the dissociation constant for the -COOH group (pKa), and the net charge index of the side chain (NCI) (detail in mSystems, 2021).
Copyright © 2022 Institute of Information Science, Academia Sinica, TAIWAN. |
All Rights reserved. |